SignalP V2.0 World Wide Web Server
SignalP predictions
Using neural networks (NN) and hidden Markov models (HMM) trained on eukaryotes
>Sequence
SignalP-NN result:
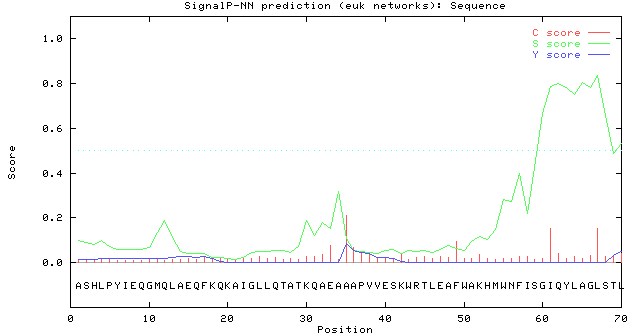
# data
>Sequence length = 70
# Measure Position Value Cutoff signal peptide?
max. C 35 0.211 0.33 NO
max. Y 35 0.086 0.32 NO
max. S 67 0.838 0.82 YES
mean S 1-34 0.082 0.47 NO
# Most likely cleavage site between pos. 34 and 35: AEA-AA
SignalP-HMM result:
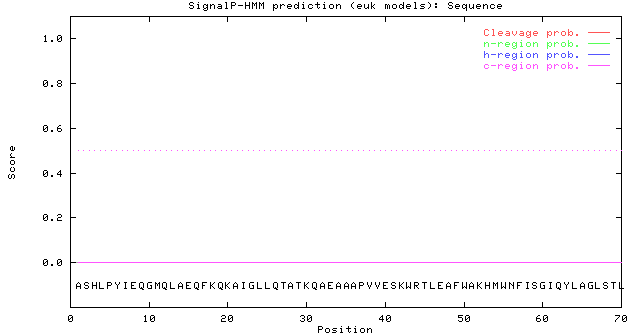
# data
>Sequence
Prediction: Non-secretory protein
Signal peptide probability: 0.003
Signal anchor probability: 0.000
Max cleavage site probability: 0.003 between pos. 34 and 35
# gnuplot
script for making the plot(s)
When using results from this server, please cite:
-
Henrik Nielsen, Jacob Engelbrecht, Søren Brunak and Gunnar von Heijne:
Identification of prokaryotic and eukaryotic signal peptides and prediction
of their cleavage sites. Protein Engineering, 10, 1-6 (1997).
If you specifically use the SignalP-HMM output, please also cite:
-
Henrik Nielsen and Anders Krogh: Prediction of signal peptides and signal
anchors by a hidden Markov model. In Proceedings of the Sixth International
Conference on Intelligent Systems for Molecular Biology (ISMB
6), AAAI Press, Menlo Park, California, pp. 122--130 (1998).


