Graphics in PostScript
# Predictions for N-Glycosylation sites in 1 sequence
Warning: This sequence may not contain a signal peptide !!
Proteins without signal peptides are unlikely to be exposed to the N-glycosylation machinery and thus may not be glycosylated (in vivo) even though they contain potential motifs. # SignalP-NN euk predictions (SignalP Output explanation) # name Cmax pos ? Ymax pos ? Smax pos ? Smean ? Sequence 0.098 32 N 0.049 32 N 0.152 13 N 0.081 N
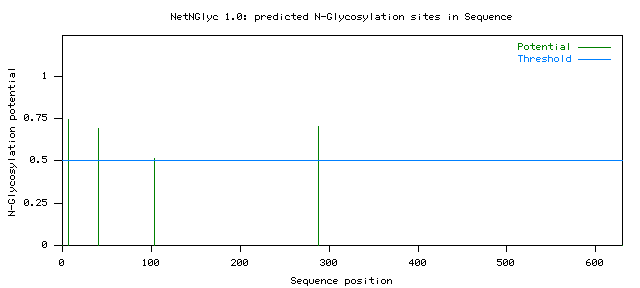
Name: Sequence Length: 630 MASLCSNSSSTSLKTPFTSSTTCLSSTPTASQLFLHGKRNKTFKVSCKVTNTNGNQDETNSVDRRNVLLGLGGLYGVANA 80 IPLAASAAPTPPPDLSSCNKPKINATTEVPYFCCAPKPDDMSKVPYYKFPSVTKLRIRPPAHALDEAYIAKYNLAISRMK 160 DLDKTQPDNPIGFKQQANIHCAYCNGGYSIDGKVLQVHNSWLFFPFHRWYLYFYERILGSLIDDPTFGLPFWNWDHPKGM 240 RFPPMFDVPGTALYDERRGDQIHNGNGIDLGYFGDQVETTQLQLMTNNLTLMYRQLVTNSPCPLMSLVDLTLFGSTVEDA 320 GTVENIPHSPVHIWVGTRRGSVLPVGKISNGEDMGNFYSAGLDPLFYCHHSNVDRMWNEWKATGGKRTDIQNKDWLNSEF 400 FFYDENGNPFKVRVRDCLDTKKMGYDYHATATPWRNFKPKTKASAGKVNTGSIPPESQVFPLAKLDKAISFSINRPASSR 480 TQQEKNAQEEVLTFNAIKYDNRDYIRFDVFLNVDNNVNANELDKAEFAGSYTSLPHVHRVGDPKHTATATLRLAITELLE 560 DIGLEDEDTIAVTLVPKKGDISIGGVEIKLAIVKLVCVVNLLTLQLNKDRFCYDSVFVCWFVCLFFNFHV ......N................................N........................................ 80 .......................N........................................................ 160 ................................................................................ 240 ...............................................N................................ 320 ................................................................................ 400 ................................................................................ 480 ................................................................................ 560 ...................................................................... (Threshold=0.5) -------------------------------------------------------------------------------- SeqName Position Potential Jury NGlyc agreement result -------------------------------------------------------------------------------- Sequence 7 NSSS 0.7437 (9/9) ++ Sequence 40 NKTF 0.6912 (9/9) ++ Sequence 104 NATT 0.5131 (6/9) + Sequence 288 NLTL 0.7029 (9/9) ++ --------------------------------------------------------------------------------