
|
|
NetPhos 2.0 Server - prediction results
Technical University of Denmark |
630 Sequence
MASLCSNSSSTSLKTPFTSSTTCLSSTPTASQLFLHGKRNKTFKVSCKVTNTNGNQDETNSVDRRNVLLGLGGLYGVANA 80
IPLAASAAPTPPPDLSSCNKPKINATTEVPYFCCAPKPDDMSKVPYYKFPSVTKLRIRPPAHALDEAYIAKYNLAISRMK 160
DLDKTQPDNPIGFKQQANIHCAYCNGGYSIDGKVLQVHNSWLFFPFHRWYLYFYERILGSLIDDPTFGLPFWNWDHPKGM 240
RFPPMFDVPGTALYDERRGDQIHNGNGIDLGYFGDQVETTQLQLMTNNLTLMYRQLVTNSPCPLMSLVDLTLFGSTVEDA 320
GTVENIPHSPVHIWVGTRRGSVLPVGKISNGEDMGNFYSAGLDPLFYCHHSNVDRMWNEWKATGGKRTDIQNKDWLNSEF 400
FFYDENGNPFKVRVRDCLDTKKMGYDYHATATPWRNFKPKTKASAGKVNTGSIPPESQVFPLAKLDKAISFSINRPASSR 480
TQQEKNAQEEVLTFNAIKYDNRDYIRFDVFLNVDNNVNANELDKAEFAGSYTSLPHVHRVGDPKHTATATLRLAITELLE 560
DIGLEDEDTIAVTLVPKKGDISIGGVEIKLAIVKLVCVVNLLTLQLNKDRFCYDSVFVCWFVCLFFNFHV 640
...........S.......S.....................T...S.................................. 80
.........T................T...Y..............Y.....................Y............ 160
...........................Y...............................S.................... 240
.................................................................S.............. 320
........S.......T...S.......S................................................... 400
..Y................T....Y......T........................S....................SS. 480
..................Y....Y.........................SY..................T.......... 560
........T............S................................................ 640
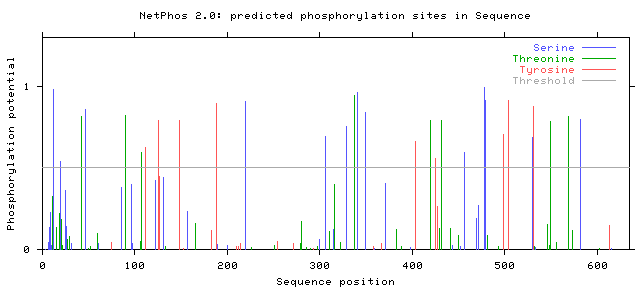
Phosphorylation sites predicted: Ser: 13 Thr: 8 Tyr: 9
Serine predictions
Name Pos Context Score Pred
_________________________v_________________
Sequence 3 --MASLCSN 0.003 .
Sequence 6 ASLCSNSSS 0.040 .
Sequence 8 LCSNSSSTS 0.135 .
Sequence 9 CSNSSSTSL 0.228 .
Sequence 10 SNSSSTSLK 0.025 .
Sequence 12 SSSTSLKTP 0.983 *S*
Sequence 19 TPFTSSTTC 0.015 .
Sequence 20 PFTSSTTCL 0.540 *S*
Sequence 25 TTCLSSTPT 0.361 .
Sequence 26 TCLSSTPTA 0.143 .
Sequence 31 TPTASQLFL 0.035 .
Sequence 46 TFKVSCKVT 0.859 *S*
Sequence 61 DETNSVDRR 0.034 .
Sequence 86 PLAASAAPT 0.378 .
Sequence 96 PPDLSSCNK 0.396 .
Sequence 97 PDLSSCNKP 0.035 .
Sequence 122 PDDMSKVPY 0.421 .
Sequence 131 YKFPSVTKL 0.444 .
Sequence 157 NLAISRMKD 0.232 .
Sequence 189 NGGYSIDGK 0.033 .
Sequence 200 QVHNSWLFF 0.023 .
Sequence 220 RILGSLIDD 0.909 *S*
Sequence 300 LVTNSPCPL 0.060 .
Sequence 306 CPLMSLVDL 0.690 *S*
Sequence 315 TLFGSTVED 0.123 .
Sequence 329 NIPHSPVHI 0.753 *S*
Sequence 341 TRRGSVLPV 0.960 *S*
Sequence 349 VGKISNGED 0.843 *S*
Sequence 359 GNFYSAGLD 0.004 .
Sequence 371 YCHHSNVDR 0.405 .
Sequence 398 DWLNSEFFF 0.013 .
Sequence 444 KTKASAGKV 0.027 .
Sequence 452 VNTGSIPPE 0.021 .
Sequence 457 IPPESQVFP 0.593 *S*
Sequence 470 DKAISFSIN 0.191 .
Sequence 472 AISFSINRP 0.270 .
Sequence 478 NRPASSRTQ 0.995 *S*
Sequence 479 RPASSRTQQ 0.912 *S*
Sequence 530 EFAGSYTSL 0.687 *S*
Sequence 533 GSYTSLPHV 0.010 .
Sequence 582 KGDISIGGV 0.796 *S*
Sequence 615 FCYDSVFVC 0.004 .
_________________________^_________________
Threonine predictions
Name Pos Context Score Pred
_________________________v_________________
Sequence 11 NSSSTSLKT 0.324 .
Sequence 15 TSLKTPFTS 0.135 .
Sequence 18 KTPFTSSTT 0.221 .
Sequence 21 FTSSTTCLS 0.185 .
Sequence 22 TSSTTCLSS 0.023 .
Sequence 27 CLSSTPTAS 0.063 .
Sequence 29 SSTPTASQL 0.079 .
Sequence 42 KRNKTFKVS 0.818 *T*
Sequence 50 SCKVTNTNG 0.009 .
Sequence 52 KVTNTNGNQ 0.021 .
Sequence 59 NQDETNSVD 0.096 .
Sequence 90 SAAPTPPPD 0.823 *T*
Sequence 106 KINATTEVP 0.049 .
Sequence 107 INATTEVPY 0.594 *T*
Sequence 133 FPSVTKLRI 0.016 .
Sequence 165 DLDKTQPDN 0.160 .
Sequence 226 IDDPTFGLP 0.014 .
Sequence 251 DVPGTALYD 0.024 .
Sequence 279 DQVETTQLQ 0.036 .
Sequence 280 QVETTQLQL 0.172 .
Sequence 286 LQLMTNNLT 0.012 .
Sequence 290 TNNLTLMYR 0.006 .
Sequence 298 RQLVTNSPC 0.017 .
Sequence 311 LVDLTLFGS 0.111 .
Sequence 316 LFGSTVEDA 0.399 .
Sequence 322 EDAGTVENI 0.041 .
Sequence 337 IWVGTRRGS 0.944 *T*
Sequence 383 EWKATGGKR 0.124 .
Sequence 388 GGKRTDIQN 0.020 .
Sequence 420 DCLDTKKMG 0.792 *T*
Sequence 430 DYHATATPW 0.129 .
Sequence 432 HATATPWRN 0.791 *T*
Sequence 441 FKPKTKASA 0.130 .
Sequence 450 GKVNTGSIP 0.084 .
Sequence 481 ASSRTQQEK 0.083 .
Sequence 493 EEVLTFNAI 0.019 .
Sequence 532 AGSYTSLPH 0.021 .
Sequence 546 DPKHTATAT 0.152 .
Sequence 548 KHTATATLR 0.027 .
Sequence 550 TATATLRLA 0.785 *T*
Sequence 556 RLAITELLE 0.040 .
Sequence 569 EDEDTIAVT 0.813 *T*
Sequence 573 TIAVTLVPK 0.116 .
Sequence 603 VNLLTLQLN 0.009 .
_________________________^_________________
Tyrosine predictions
Name Pos Context Score Pred
_________________________v_________________
Sequence 75 LGGLYGVAN 0.042 .
Sequence 111 TEVPYFCCA 0.627 *Y*
Sequence 126 SKVPYYKFP 0.790 *Y*
Sequence 127 KVPYYKFPS 0.449 .
Sequence 148 LDEAYIAKY 0.788 *Y*
Sequence 152 YIAKYNLAI 0.009 .
Sequence 183 IHCAYCNGG 0.117 .
Sequence 188 CNGGYSIDG 0.895 *Y*
Sequence 210 FHRWYLYFY 0.019 .
Sequence 212 RWYLYFYER 0.020 .
Sequence 214 YLYFYERIL 0.039 .
Sequence 254 GTALYDERR 0.049 .
Sequence 272 IDLGYFGDQ 0.034 .
Sequence 293 LTLMYRQLV 0.007 .
Sequence 358 MGNFYSAGL 0.021 .
Sequence 367 DPLFYCHHS 0.034 .
Sequence 403 EFFFYDENG 0.661 *Y*
Sequence 425 KKMGYDYHA 0.557 *Y*
Sequence 427 MGYDYHATA 0.262 .
Sequence 499 NAIKYDNRD 0.703 *Y*
Sequence 504 DNRDYIRFD 0.914 *Y*
Sequence 531 FAGSYTSLP 0.875 *Y*
Sequence 613 DRFCYDSVF 0.148 .
_________________________^_________________
 Explain the output. Go back.
Explain the output. Go back.


 Explain the output. Go back.
Explain the output. Go back.