 Why
is it important to load equal amounts of mRNA?
Why
is it important to load equal amounts of mRNA? Why
is it important to load equal amounts of mRNA?
Why
is it important to load equal amounts of mRNA?|
First, let us focus on GAPDH results (lower panel of the figure to the right).  Is
there a difference in GAPDH expression between the three tissues compared? Is
there a difference in GAPDH expression between the three tissues compared?
|
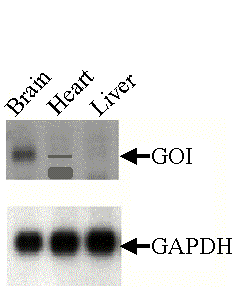 |
| Now, let us focus
on the GOI results (upper panel of
the figure to the right).
|
 |
|
|
|
One of the uses of Northern blots is to compare mRNA quantities in different samples. Loading samples containing unequal amounts of mRNA will bias the results and will lead to wrong interpretations of the data.
.