The Northern Blot method reveals information about RNA identity, size and abundance.
|
|
|
The Northern Blot procedure is quite similar to that of Southern Blot, except that the biomaterial used is RNA instead of DNA.
Letís review the procedure step by step:
| 1 | RNA is isolated
from several biological samples (e.g. various tissues, various developmental
stages of same tissue etc.)
* RNA is more susceptible to degradation than DNA. * The decision whether to isolate total RNA or mRNA depends on the scientific question at hand. mRNA comprises 2-5% of total RNA, and its isolation requires a further purification step using a poly A tail hybridization. |
|
| 2 | The RNA samples are separated according to their size on an agarose gel | Sampleas are loaded on gel 
The resulting gel following the run  |
| 3 | The gel is then blotted on a nylon membrane or a nitrocellulose filter. | 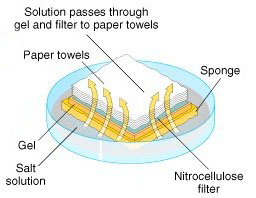 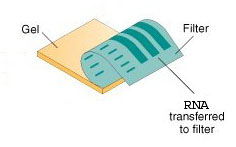 |
| 4 | The membrane is placed in a dish containing hybridization buffer with a labeled probe (radioactively or chemically labeled). The probe is specifically designed for the sequence of interest. Thus, it will hybridize to the RNA on the blot that corresponds to the sequence of interest. |  The
probe can be either ss-DNA or RNA. The
probe can be either ss-DNA or RNA. |
| 5 | The membrane is washed to remove unbound probe. |  Access
probe is removed Access
probe is removed |
| 6 | The labeled probe
is detected via autoradiography (if a radioactive probe is
used) or via a chemiluminescence reaction (if a chemically
labeled probe is used). In both cases this results
in the formation of a dark band on an X-ray film.
We can now compare expression patterns of the sequence of interest in the different samples. |
 |
Improvements in sensitivity
of this method can be achieved by using high specific activity antisense
RNA probes (step 4), optimized hybridization buffers (step 4) and positively
charged nylon membranes (step 3).
|
|
|
| Related Protocols: | Troubleshooting(scroll down to bottom of page) |
|
|
|
|
|
|
|
|
|
|
.