

2. Possible but unlikely - usually the incorporation is not that efficient. Possibly, you forgot to add X gal and or IPTG to the plate, and so you cannot distinguish between positive and negative results.

3. Wrong - not adding all the antibiotics should increase the number of surviving colonies. The lack of colonies may be due to killing the cells at the time of electroporation during bacterial transformation. Alternatively, you have added too much antibiotics.

4. Right
- you should now isolate and characterize some of the white colonies,
since they should contain the recombinant viral genome with the BeHappyReceptor
gene (![]() ).
).
|
|
 |
|
In order to screen for the insertion of our gene into
the viral genome, you now conduct a Colony PCR assay for one colony from
each streak (
|
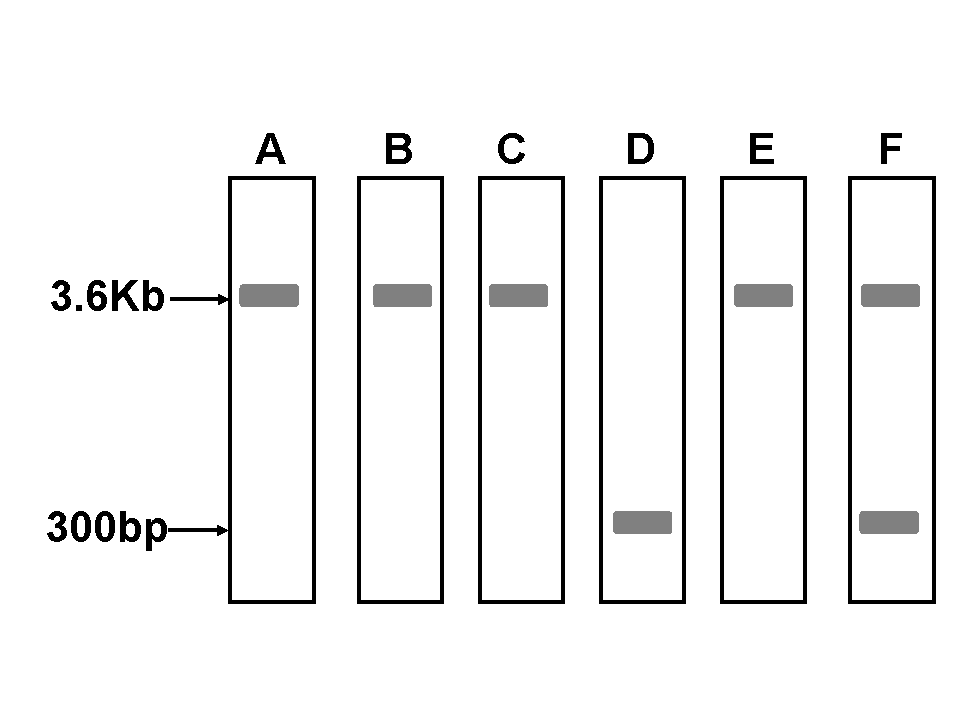 |
A. Colony A B. Colony F C. Colony D. |
A. Correct, but this is not the most accurate answer,

B. Wrong - colony F contains a hetrogenous population of both recombinant and native viral genomes.

C. Wrong - although the original colony appeared white, this is a homogenous population of bacterial cells that did not undergo transposition and ontains the native LacZ-undisrupted viral genome. Sometimes, when the density of the original colonies is too high, the local concentration of the Xgal substrate in the plate is depleted and is insufficient to induce the expected blue appearance of this colony.

D. Right - each of these colonies represents a successful event of transposition. You may safely use each of these colonies for DNA preparation to be used at the next step - viral DNA transfection into SF9 insect cells for production of Baculovirus.

-